Graphical Abstract
RNA m6A modification is the most common modification of mRNA in eukaryotes. In recent years, it has been involved in different biological processes and played a key regulatory role in species such as human, animal, plant and yeast. In these species, METTL3/IME4 is mainly in charge of catalyzing the m6A modification of RNA molecules on N6-methyladenine, and is one of the most important constituent proteins of the m6A methyltransferase complex. However, since no protein homologous to METTL3/IME4 has been found in filamentous fungi, whether m6A modification exists and plays a role in phytopathogenic fungi needs further research. The authors of this study identified MTA1 (generally thought to encode a key enzyme in DNA 6mA modification) homologous to the human METTL4 protein and subsequently demonstrated that this protein is involved in R. oryzae RNA m6A modification, but whether it is involved in DNA 6mA modification requires further investigation).
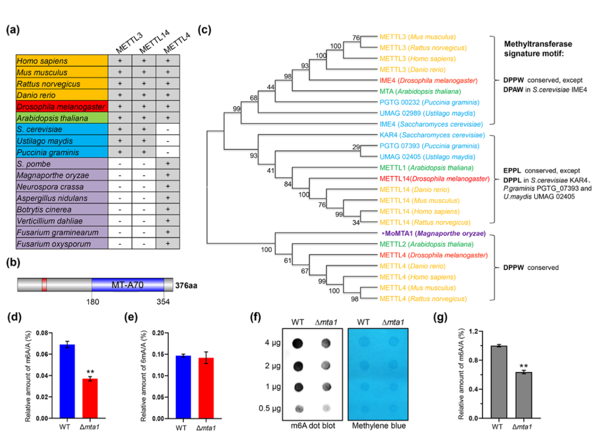 Fig 1. MTA1 is involved in M. oryzae RNA m6A modification. (New Phytologist, 2022)
Fig 1. MTA1 is involved in M. oryzae RNA m6A modification. (New Phytologist, 2022)
With MTA1 gene knockout, the total m6A modification of oryzae was significantly reduced, and the pathogenicity of the bacteria was seriously reduced. The reduced virulence analysis found that the function of the appressorium of the knockout body was affected, and the ability of infecting hyphae to expand was also inhibited. Knockout of appressorium formation resulted in significantly restricted glycogen and liposome utilization, blocked autophagy, and significantly reduced appressorium pressure. Since MTA1 plays a key role in the appressorium, the authors collected the appressorium material from wild-type and MTA1 knockout bodies for combined MeRIP-seq and RNAseq analysis. MeRIP seq analysis identified 595 mRNAs with reduced m6A methylation at 659 sites in the appressorium of the ∆mta1 strain compared with the wild-type strain. Among them, carbohydrate metabolism and lipid metabolism, and autophagy-related signaling pathways were enriched. Combined with RNAseq data, 114 m6A modifications were found to be negatively correlated with mRNA abundance, suggesting that m6A modifications may be involved in mRNA degradation.
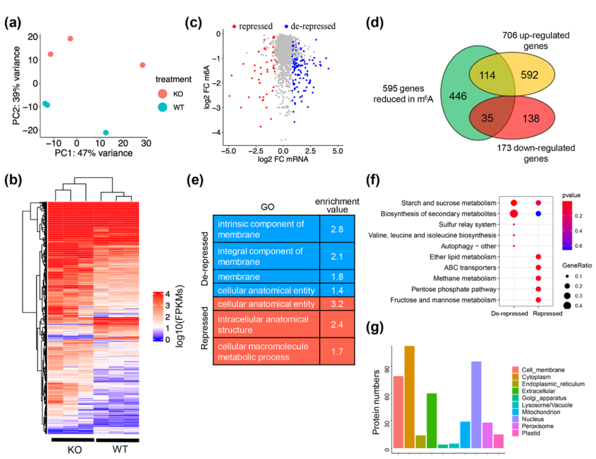 Fig 2. Combined m6A-seq and RNA-seq analysis. (New Phytologist, 2022)
Fig 2. Combined m6A-seq and RNA-seq analysis. (New Phytologist, 2022)
Further analysis revealed that the transcripts of some ATG genes regulating autophagy were modified by m6A, including ATG8. The data predicted that the A982 site in the 3' untranslated region of the ATG8 transcript might be the m6A modification site, and then site mutation was performed on this site. MeRIP-qPCR analysis showed that the A982 site mutation caused serious decline in the level of ATG8 m6A modification; meanwhile, qRT-PCR analysis showed that the A982 site mutation led to increased the level of ATG8 mRNA, indicating that m6A negatively regulates the mRNA abundance of ATG8. After the A982 site mutation, the level of ATG8 protein is increased at the same time, and the autophagy process of appressorium cells is disturbed, thereby affecting the function of the appressorium and reducing the pathogenicity. Based on the above studies, it was found that in the process of appressorium formation in rice blast, m6A modification may be involved in the regulation of autophagy protein mRNA abundance, thereby coordinating the autophagy of appressorium cells, regulating the formation of functional appressorium, and helping rice blast fungus disease.
The latest molecular plant report reveals the molecular mechanism of rice redox sensor in signal transduction of osmotic stress, provides novel ideas and foundation for rice research and development. Creative Biolabs offers model organism antibody custom service. Just contact us to choose the right anti rice bZIP68 or anti rice GPX1 products.
The work published in the well-known scientific journals "New Phytologist".
Story Source: Ren Z, Tang B, Xing J, et al. MTA1‐mediated RNA m6A modification regulates autophagy and is required for infection of the rice blast fungus[J]. New Phytologist, 2022.
Creative Biolabs is committed to provide high-quality reagents and one-stop technical services for model organism researchers worldwide. We have a full line of high-quality antibodies targeting to rice m6A and Yeast m6A. Please feel free to contact us.